Motivation¶
Despite its numerous advantages, Newton’s method suffers from two disadvantages:
Computing Newton’s step involves calculating and inverting the Hessian matrix , which becomes very costly in high dimensions.
The Hessian matrix is not always positive definite, so it’s not always possible to calculate its inverse.
Solution: We replace the Hessian matrix with an approximation , leading to so-called quasi-Newton methods.
Principle of Quasi-Newton Methods¶
The crucial step lies in defining matrices (or ) such that
often by a recursive relation (i.e., it is built on the fly, from values at previous iterations). For a given update rule, one gets a specific quasi-Newton method. Some examples are:
Broyden-Fletcher-Goldfarb-Shanno (BFGS), probably the most popular
L-BFGS: memory-efficient BFGS
Symmetric rank-one (SR1)
Davidon–Fletcher–Powell (DFP), etc.
Many of such algorithms are implemented in standard Python libraries, such as in scipy. Check for instance the documentation of scipy.optimize.minimize function.
Example: BFGS algorithm¶
For details about the derivation of BFGS updates, properties of the algorithm and implementation details, one can consult Section 6.1, Nocedal & Wright, 2006.
Comparison on a non-quadratic example¶
To compare the BFGS algorithm with gradient descent (fixed step-size) and Newton’s method, we come back to the non-quadratic example introduced when studying step-size strategies for gradient descent
Let us look at how fixed step-size gradient descent and Newton’s method perform on this problem.
Source
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import minimize
def f(x1, x2):
return np.exp(x1 +3*x2 - 0.1) + np.exp(x1 -3*x2- 0.1) + np.exp(-x1 - 0.1)
def dfdx(x1, x2):
dx1 = np.exp(x1 + 3*x2 - 0.1) + np.exp(x1 - 3*x2 - 0.1) - np.exp(-x1 - 0.1)
dx2 = 3 * np.exp(x1 + 3*x2 - 0.1) - 3 * np.exp(x1 - 3*x2 - 0.1)
return np.array([dx1, dx2])
def d2fd2x(x1, x2):
hess = np.array([
[
np.exp(x1 + 3*x2 - 0.1) + np.exp(x1 - 3*x2 - 0.1) + np.exp(-x1 - 0.1),
3*np.exp(x1 + 3*x2 - 0.1) - 3*np.exp(x1 - 3*x2 - 0.1)
],
[
3*np.exp(x1 + 3*x2 - 0.1) - 3*np.exp(x1 - 3*x2 - 0.1),
9*np.exp(x1 + 3*x2 - 0.1) + 9*np.exp(x1 - 3*x2 - 0.1)
]
])
return hess
def gradient_descent(xinit, dfdx, alpha, niter=10):
xn = np.zeros((2, niter + 1))
xn[:, 0] = xinit
for n in range(niter):
grad = dfdx(xn[0, n], xn[1, n])
xn[:, n + 1] = xn[:, n] - alpha * grad
return xn
def newton_method(xinit, dfdx, d2fd2x, niter=10):
xn = np.zeros((2, niter + 1))
xn[:, 0] = xinit
for n in range(niter):
grad = dfdx(xn[0, n], xn[1, n])
hess = d2fd2x(xn[0, n], xn[1, n])
xn[:, n + 1] = xn[:, n] - np.linalg.pinv(hess) @ grad
return xn
def plot_path(xs, label):
xs = np.array(xs)
plt.plot(xs[0,:], xs[1,:], marker='o', label=label)
# starting point
xinit = [-2, .5]
alpha = 0.1
xn_grad = gradient_descent(xinit, dfdx, alpha, niter=50)
xn_newton = newton_method(xinit, dfdx, d2fd2x, niter=20)
# plot
x1s = np.linspace(-2, 0.6, 50)
x2s = np.linspace(-0.5, 0.5, 50)
X1, X2 = np.meshgrid(x1s, x2s)
Z = f(X1, X2)
plt.figure(figsize=(8, 6))
cp = plt.contourf(X1, X2, Z, levels=30, cmap='viridis')
plot_path(xn_grad, r"Fixed step-size $\alpha$" + f"$= {alpha}$")
plot_path(xn_newton, "Newton's method")
plt.legend()
plt.colorbar(cp)
plt.xlabel("$x_1$")
plt.ylabel("$x_2$")
plt.title("$f_0$")
plt.show()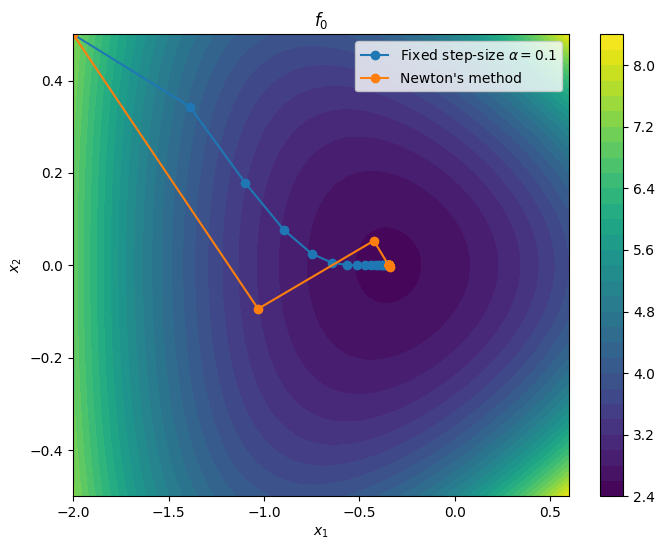
Below is a possible, non-optimized, BFGS implementation.
def BFGS(xinit, dfdx, Hinit, alphainit, f, niter=10, s=0.3, eta=0.9):
xn = np.zeros((2, niter + 1))
xn[:, 0] = xinit
Hk = Hinit
grad = dfdx(xn[0, 0], xn[1, 0])
for n in range(niter):
alpha = alphainit
xtemp = xn[:, n] - alpha * Hk @ grad
# check backtracking condition
ftemp = f(xtemp[0], xtemp[1])
backtrack_comp = f(xn[0, n], xn[1, n]) - alpha * s * grad.T @ Hk @ grad
while ftemp > backtrack_comp:
alpha = eta * alpha
xtemp = xn[:, n] - alpha * Hk @ grad
ftemp = f(xtemp[0], xtemp[1])
backtrack_comp = f(xn[0, n], xn[1, n]) - alpha * s * grad.T @ Hk @ grad
xn[:, n + 1] = xtemp
# update Hk
if n < niter - 1:
newgrad = dfdx(xn[0, n + 1], xn[1, n + 1])
sk = xn[:, n + 1] - xn[:, n]
yk = newgrad - grad
rhok = 1.0 / (yk.T @ sk)
grad = newgrad
I = np.eye(2)
mat = I - rhok * np.outer(sk, yk)
mat2 = I - rhok * np.outer(yk, sk)
Hk = mat @ Hk @ mat2 + rhok * np.outer(sk, sk)
return xnLet us compare how BFGS performs, starting from the same initial point and using .
Source
# BFGS
Hinit = np.linalg.pinv(d2fd2x(xinit[0], xinit[1]))
xn_BFGS = BFGS(xinit, dfdx, Hinit, 1, f, niter=10, s=0.3, eta=0.5)
# plot
x1s = np.linspace(-2, 0.6, 50)
x2s = np.linspace(-0.5, 0.5, 50)
X1, X2 = np.meshgrid(x1s, x2s)
Z = f(X1, X2)
plt.figure(figsize=(8, 6))
cp = plt.contourf(X1, X2, Z, levels=30, cmap='viridis')
plot_path(xn_grad, r"GD with fixed step-size $\alpha$" + f"$= {alpha}$")
plot_path(xn_newton, "Newton's method")
plot_path(xn_BFGS, "BFGS")
plt.legend()
plt.colorbar(cp)
plt.xlabel("$x_1$")
plt.ylabel("$x_2$")
plt.title("$f_0$")
plt.show()
# compute optimal value
res = minimize(lambda x: f(x[0], x[1]), xinit)
pstar = res.fun
# Plot objective values for each method
plt.figure(figsize=(8, 6))
plt.semilogy([f(*x) - pstar for x in xn_grad.T], label=r"GD with fixed step-size $\alpha$" + f"$= {alpha}$", marker='o')
plt.semilogy([f(*x)- pstar for x in xn_newton.T], label="Newton's method", marker='o')
plt.semilogy([f(*x)- pstar for x in xn_BFGS.T], label=f"BFGS", marker='o')
plt.xlabel("Iteration")
plt.ylabel("$f_0(x) - p^\star$")
plt.title("suboptimality value vs iteration")
plt.legend()
plt.grid(True)
plt.show()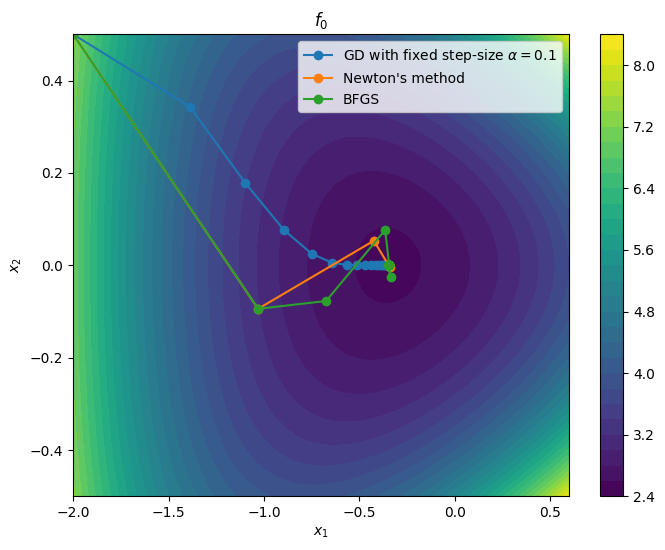
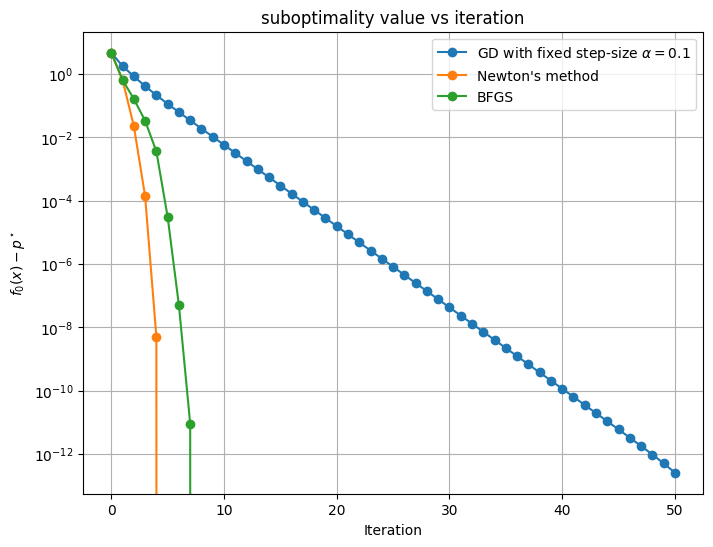
In this example, we see that BFGS does converges almost as fast as Newton’s method -- and much faster than fixed step-size gradient descent. However, unlike Newton’s method, it does not require the evaluation of the Hessian at each iteration.
- Nocedal, J., & Wright, S. J. (2006). Numerical optimization (Second Edition). Springer.