The main hypothesis is that there exists a linear relationship between data and variables .
Figure 1:illustration of the linear model in the overdetermined case ()
Definitions and Notations¶
The vector contains the data, also named observations or measurements, which we want to model;
The matrix encodes the model (assumed known).
The vector represents the unknowns.
The vector expresses errors (model, noise, etc.).
For simplicity, we assume that observations are linearly independent. This is equivalent to saying that observations are not redundant. The problem (1) is said to be
overdetermined when (more observations than unknowns)
underdetermined when (strictly less observations than unknowns)
The over/under determination property play an important role regarding the uniqueness of solutions to the least squares problem, as we will see later on.
Interpretation as an unconstrained quadratic program (QP)¶
The objective function can be rewritten as
such that the least-squares optimization problem can be equivalently reformulated as the unconstrained quadratic program
where and .
Examples¶
Polynomial regression¶
Assume we are given some data as a function of time , as shown below:
Source
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(2025)
def f(x):
return x**3 + 4*x**2 - x + 1
N = 20
ts = np.linspace(-4., 2., N)
y = f(ts) + np.random.randn(N)
plt.scatter(ts, y, label="data $y$")
plt.xlabel(r"$t$")
plt.ylabel(r"$y$")
plt.legend()
plt.show()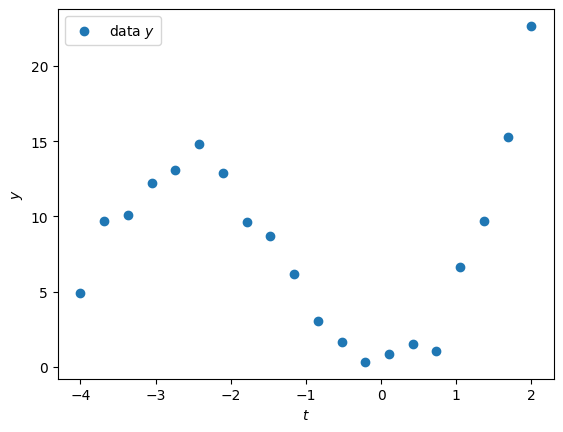
The trend clearly looks polynomial. If we assume the degree of the polynomial is known (here ), we can write a linear model to fit the data . The approach very classical: hence it is implemented in many libraries, such as scikit-learn (see PolynomialFeatures and LinearRegression).
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
# construct polynomial regression matrix
degree = 3
poly = PolynomialFeatures(degree)
X_poly = poly.fit_transform(ts.reshape(-1, 1))
# fit the data (solve the LS problem)
model = LinearRegression(fit_intercept=False)
model.fit(X_poly, y)
y_pred = model.predict(X_poly)
# display results
plt.scatter(ts, y, label="data $y$")
plt.plot(ts, f(ts), color="green", linestyle="--", label="ground truth")
plt.plot(ts, y_pred, color="red", label=f"degree {degree} fit")
plt.xlabel("$t$")
plt.ylabel("$y$")
plt.legend()
plt.show()
# Print the estimated polynomial coefficients
print("Estimated coefficients (in increasing powers of t):\n", model.coef_)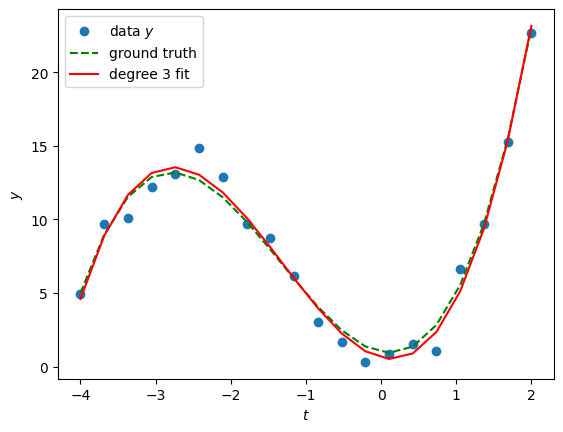
Estimated coefficients (in increasing powers of t):
[ 0.61187122 -1.2593056 4.15376347 1.05475994]
In TP1, we will see what’s under the hood of such tools and implement the solution from scratch.
2D image deconvolution¶
Assume we observe a blurred image , which is the result of convolving a true image with a known kernel , plus noise :
where denotes 2D convolution.
Example
from skimage import data
from scipy.signal import convolve2d
import matplotlib.pyplot as plt
import numpy as np
# Load cameraman image (grayscale) and decimitaed
X = data.camera()[::3, ::3]
# Define blur kernel
K = np.array([[1, 2, 1],
[2, 4, 2],
[1, 2, 1]], dtype=float)
K /= K.sum()
# Convolve image with kernel
Y = convolve2d(X, K, mode='same', boundary='symm')
# Display original and blurred images side by side
fig, axs = plt.subplots(1, 2, figsize=(8, 4))
axs[0].imshow(X, cmap='gray')
axs[0].set_title("Original image $X$")
axs[0].axis('off')
axs[1].imshow(Y, cmap='gray')
axs[1].set_title("Blurred image $Y$")
axs[1].axis('off')
plt.tight_layout()
plt.show()
We can rewrite the convolution as a linear operation (see also below) using the vectorization operator . Define , and , the above observation model is rewritten as
where is the matrix representing convolution with . The least squares problem is then:
Expressing 2D convolution as a matrix–vector operation
A 2D discrete convolution between an image and a kernel is defined as
where , , and appropriate boundary conditions (e.g. zero-padding) are assumed.
Matrix–vector Form
Flatten the image into a vector using row-major ordering (or column-major ordering).
Construct the convolution matrix .
Each row of corresponds to one output pixel.
The kernel values are placed in the columns corresponding to the input pixel locations that contribute to that output.
Formally, if we index pixels by a single index , then
This means is a block Toeplitz matrix with Toeplitz blocks (BTTB). For an image of height and width :
where each block is itself a Toeplitz matrix encoding horizontal shifts of the kernel.
The convolution becomes
where is the flattened output image.
For the above example, is a huge structured matrix of size . To simplify the presentation, assume (the matrix is already !)
Source
import matplotlib.patches as patches
def build_convolution_matrix(kernel, image_shape):
"""
Build a convolution matrix A such that:
y_flat = A @ x_flat
where x_flat is the flattened image, and kernel is the convolution filter.
"""
H, W = image_shape
kH, kW = kernel.shape
pad_h, pad_w = kH // 2, kW // 2
# Number of pixels
N = H * W
# Initialize the convolution matrix
A = np.zeros((N, N))
# Iterate over each pixel in the image
for i in range(H):
for j in range(W):
row = i * W + j # Row index in A
# Place kernel centered at (i,j)
for m in range(kH):
for n in range(kW):
ii = i + m - pad_h
jj = j + n - pad_w
if 0 <= ii < H and 0 <= jj < W:
col = ii * W + jj
A[row, col] = kernel[m, n]
return A
image_shape = 10, 10
A = build_convolution_matrix(K, image_shape)
fig, ax = plt.subplots(figsize=(8,8))
im = ax.imshow(A, cmap='gray_r', interpolation='none')
plt.title("Full Convolution Matrix $A$")
plt.colorbar(im, ax=ax, fraction=0.046, pad=0.04)
# Inset: show top-left 2 blocks along each direction
block_size_x = image_shape[0]
block_size_y = image_shape[1]
inset_size = block_size_x + block_size_y
# Create inset axes
ax_inset = ax.inset_axes([0.6, 0.6, 0.35, 0.35])
ax_inset.imshow(A[:20, :20], cmap='gray_r', interpolation='none')
ax_inset.set_title('first two blocks of $A$')
plt.show()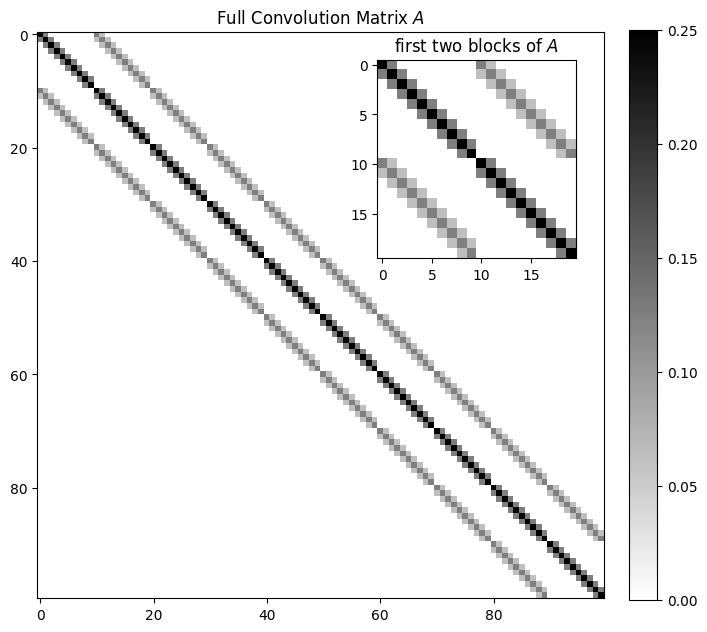
In practice, due to the large size of the matrix , we do not use direct solutions based on pseudo-inverse calculations (see later on). In this case, the least squares problem is solved through the use of FFTs.
from numpy.fft import fft2, ifft2
# compute least squares solution using FFT
Y_fft = fft2(Y)
K_fft = fft2(K, s=X.shape)
eps = 1e-6*np.max(np.abs(K_fft))
X_hat = np.real(ifft2(Y_fft/(1+K_fft+eps))) # small epsilon for stability
# Display results
fig, axs = plt.subplots(1, 3, figsize=(10, 3))
axs[0].imshow(X, cmap='gray')
axs[0].set_title("True image $X$")
axs[1].imshow(Y, cmap='gray')
axs[1].set_title("Blurred observation $Y$")
axs[2].imshow(X_hat, cmap='gray')
axs[2].set_title("LS estimate $\hat{X}$")
for ax in axs: ax.axis('off')
plt.tight_layout()
plt.show()
This example demonstrates how 2D deconvolution can be formulated and solved as a linear least squares problem.